About NCVC
Researchers identify a critical inhibitory site in an energy-producing enzyme, which can lead to rational design of antibiotics.
Antimicrobial resistance (AMR) is a global health problem. Handful species of drug-resistant pathogens, including Neisseria gonorrhoeae, could become untreatable due to a high degree of AMR. A team of researchers has discovered a new mechanism to allosterically inhibit the critical enzyme in the respiratory chain, widely conserved in all three domains of life: bacteria, archaea, and eukaryotes. The team has succeeded in identifying an antibiotic based on their findings, which is effective against a super drug-resistant strain of Neisseria gonorrhoeae.
The team published their findings in the journal Nature Communications.
Antimicrobial resistance (AMR) is a global health problem. Many efforts have been made to reduce the burden of AMR perils globally since 2013 (Figure 1A). Yet, threats from some species continue to rise regardless: drug-resistant Neisseria gonorrhoeae is one of five urgent threats. Resistance to ceftriaxone, the last option for an empirical first-line antibiotic against Neisseria gonorrhoeae in most countries, has been reported and continues to emerge globally. The gonococcal infection could become untreatable due to a high degree of AMR, which would increase serious complications: infertility, ectopic pregnancy, and increased transmission of HIV, and neonatal keratoconjunctivitis that can lead to blindness. The emergence of resistant pathogens to currently available antibiotics is very alarming; thus, the development of antibiotics with a novel mechanism of action is seriously required.
The respiratory chain has recently garnered considerable scientific attention as a potential target for antibiotics. As respiratory enzymes are essential for life, their core structure is generally conserved from bacteria to mammals. Thus, the surface of the substrate binding pocket is quite similar among species, making it challenging to develop a competitive inhibitor for the substrate binding pocket. Another type of inhibitor for an enzyme is an allosteric inhibitor. Another type of enzyme inhibitor, an allosteric inhibitor, causes a structural change of the enzyme, leading to the inhibition of its activity. Allosteric sites are evolutionarily less conserved in amino acid sequence than substrate binding sites, theoretically improving selectivity and reducing toxicity. However, a systematic and strategic search for allosteric inhibitors has yet to be established.
The team identified an allosteric inhibitory site buried inside mammalian mitochondrial heme-copper oxidases (mtHCOs), the essential respiratory enzymes for life. The steric conformation around the binding pocket of HCOs is highly conserved among bacteria and mammals, yet the latter has an extra helix (Figure 1B, C). The existence of an additional helix in mammalian mtHcO makes surface of the pockets distinct from bacterial HCOs (Figure 1D, E). Thus, fitting inhibitors to each corresponding site must have a different character/profile. This structural difference in the conserved allostery enabled us to rationally identify bacterial HCO-specific inhibitors: an antibiotic compound, Q275, against ceftriaxone-resistant Neisseria gonorrhoeae (Figure 1F).
Future perspective for finding allosteric modulators
Our approach can be applied to finding allosteric modulators in other therapeutic targets. Enzymes generally acquire additional domains or subunits along molecular evolution, bigger in eukaryotes than in their bacterial counterparts. They could likely contain allostery inside the protein at the boundary of the structures between eukaryotes and bacteria, leading to the development of novel antibiotics, as the respiratory chain is a proven target for antibiotics. Furthermore, any fundamental molecule essential for life and conserved among species could be a potential target. Also, the additional peptides might contain a positive allosteric site at the border of their core structure; a positive allosteric modulator for the loss-of-function human disease could be a therapeutic direction. Thus, in conclusion, this study will open new avenues in protein science and therapeutic development, especially for antibiotics with a novel mechanism of action.
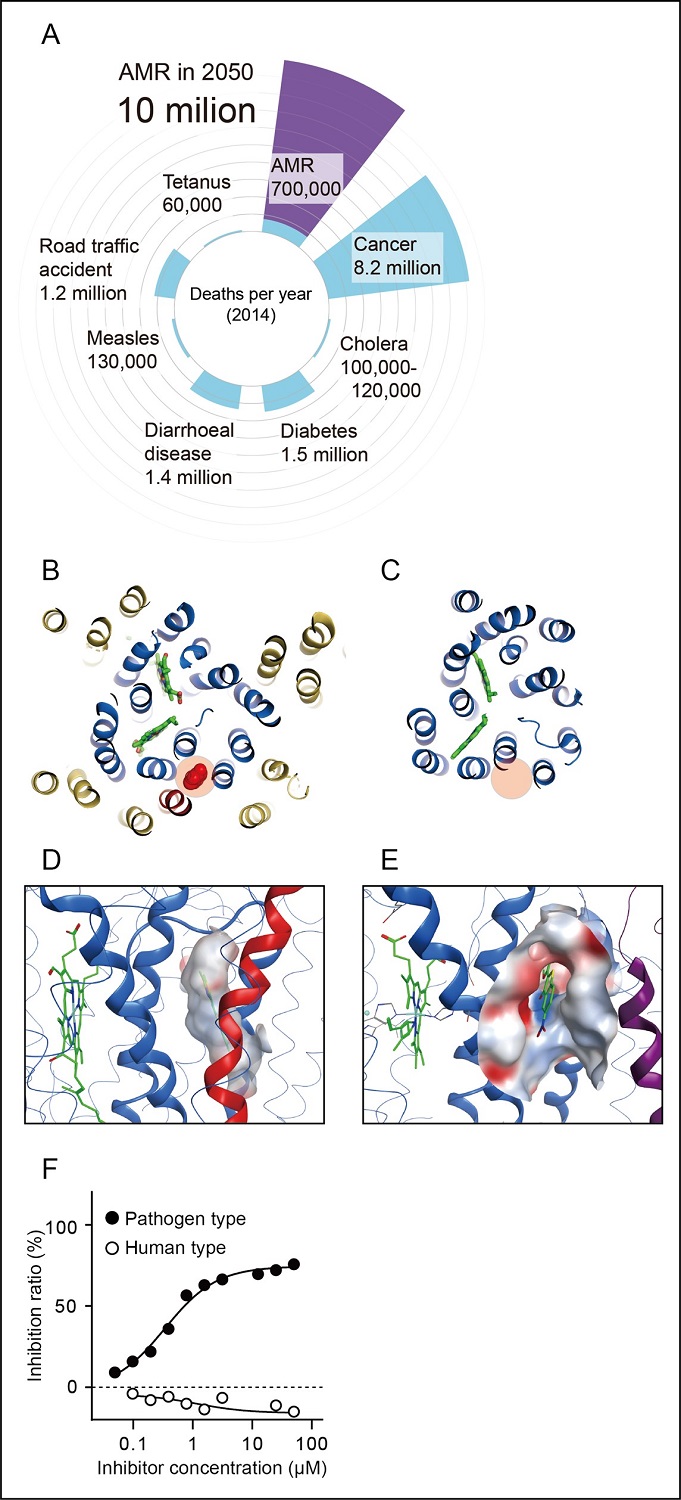
Figure 1. (A) Deaths from AMR are predicted to rise globally in the next 35 years. (O’Neill J, Chair. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London, UK: Review on Antimicrobial Resistance; 2016; p. 1-84.) (B, C) Allosteric sites (red shadow) of mammalian mtHCO (A: PDB 7XMB) and bacterial HCO (B: PDB 6FWF). Helices of subunit I are shown as blue, and a mammalian-specific subunit, COX7C (shown as red), covers the surface of the inhibitor. The other helices are shown in yellow. (D, E) The surface of the allosteric site is distinct from each other (C: mtHCO, D: bacterial HCO), which makes fitting inhibitors to each corresponding site must have a different character/profile. (F) Q275 is a specific Neisseria HCO inhibitor (filled circle), did not inhibit mammalian mtHCO (open circle).
Paper information
Yuya Nishida, Sachiko Yanagisawa, Rikuri Morita, Hideki Shigematsu, Kyoko Shinzawa-Itoh, Hitomi Yuki, Satoshi Ogasawara, Ken Shimuta, Takashi Iwamoto, Chisa Nakabayashi, Waka Matsumura, Hisakazu Kato, Chai Gopalasingam, Takemasa Nagao, Tasneem Qaqorh, Yusuke Takahashi, Satoru Yamazaki, Katsumasa Kamiya, Ryuhei Harada, Nobuhiro Mizuno, Hideyuki Takahashi, Yukihiro Akeda, Makoto Ohnishi, Yoshikazu Ishii, Takashi Kumasaka, Takeshi Murata, Kazumasa Muramoto, Takehiko Tosha, Yoshitsugu Shiro, Teruki Honma, Yasuteru Shigeta, Minoru Kubo, Seiji Takashima, Yasunori Shintani
Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases
Nature communications
This work was supported by funding from
AMED under Grant Number, JP19im0210617, JP21ek0109499 and JP22fk0108632.
Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science under Grant Numbers, 21H02914, 21K15443
Japan Science and Technology Agency-Core Research for Evolutional Science and Technology (CREST) under Grant Number, JPMJCR14M2
last updated:2022/12/09
