Research Institute
Laboratory of Signal Transduction
We are looking at naturally occurring, secretory peptides. What for?
In most areas of life science, peptides, defined as smaller (< 10,000 Da) polypeptides, have long been considered an accumulation of protein degradation products. However, some of them are functional in an organism and collectively referred to as biologically active peptides. They include peptide hormones, neuropeptides, antimicrobial peptides and venom peptides, being produced from translated proteins as a result of specific proteolytic processing.
Technological advances in mass spectrometry, along with bioinformatics, have allowed us to acquire an abundance of peptide sequences on a chromatographic time scale. By examining peptides found in the supernatant of cultured cells, we have found a regularity in the peptide landscape that had remained largely unknown until the advent of peptidomics, a subdiscipline of proteomics (for an excellent review, see Schrader M, Schultz-Knappe P and Fricker LD. EuPA Open Proteomics, 3: 171-82, 2014).
The major focus of our research is to identify novel bioactive peptides using a mass spectrometry platform. We are particularly interested in peptide hormones and neuropeptides; they are a class of cell-to-cell signaling molecules produced in vivo from translated proteins. Once a novel bioactive peptide is identified, its impact ranges from basic signaling mechanisms through the potential implication in physiology and pathophysiology. Bioactive peptides constitute an integral part of humoral regulators also in the cardiovascular system. They are known as a promising target for drug discovery and development, since they do not penetrate the cell membrane or interact with/perturb genetic material to cause unexpected adversary effects. In fact, drugs used in clinics target peptides themselves, their receptors, and peptide processing enzymes. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) has proved a robust and high-throughput method for peptide identification. We have advocated a streamlined approach to identify bioactive peptide candidates (Figure), which led to the discovery of six bioactive peptides so far, including neuroendocrine regulatory peptides (NERPs).
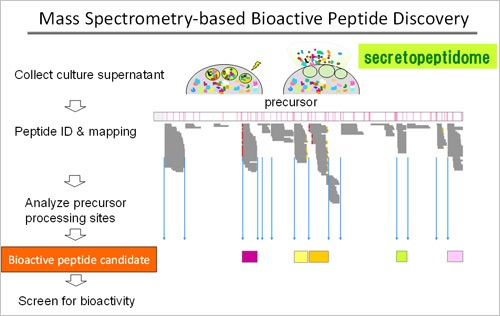
1. Mass spectrometry-based profiling of secretory peptides for bioactive peptide discovery
In general, a conventional workflow for peptide discovery involves an activity-guided biochemical purification using a specific bioassay system, an acquisition of partial amino acid sequences, and subsequent cDNA cloning. Alternatively, we have implemented a "sequence-first"approach by profiling peptides secreted by cells in culture (Figure). The information obtained by LC-MS/MS can be used to identify bioactive peptide candidates, which are subject to subsequent studies to test for their bioactivity. Given that most bioactive peptides are extracellularly released, we applied an exocytosis stimulus to cultured cells of endocrine origin to harvest liberated peptides. In so doing, we can boost the regulated secretory pathway to let the cells secrete an abundance of peptides quickly. This protocol allowed us to identify novel amidated peptides, designated neuroendocrine regulatory peptides, from the neurosecretory protein VGF (J Biol Chem 282: 26354-60, 2007). They are expressed by a defined set of endocrine cells and hypothalamic neurons. We also succeeded in identifying an intramolecular disulfide-linked 22-residue amidated peptide. It arises from insulin like growth factor-binding protein 5 (J Proteome Res 10: 1870-80, 2011). Of note, these peptides are flanked at both sides by consensus cleavage motifs of the processing enzymes responsible for peptide hormone or neuropeptide processing. The endogenous molecular form in normal tissues used to be verified with conventional methods using antibodies, but improved mass spectrometry techniques have now even dispensed with antibodies and permitted the determination straightforwardly and exactly. Of note, they are identical to those originally found in culture medium.
2. Peptidomics for studying limited proteolysis
Peptides/proteins are secreted via the regulated or constitutive secretory pathways. In cell culture models, peptides released via the regulated pathway are harvested by applying a quick exocytosis stimulus. However, most cells in our body are devoid of secretory granules and only equipped with the constitutive pathway. This means that secretory material is difficult to retrieve efficiently, without compromising the integrity of peptide constituents. Using rat primary cultured cardiac fibroblasts as a model system, we have demonstrated that peptide profiling highlights limited proteolysis (specific cleavage) sites of secretory proteins or transmembrane proteins. Limited proteolysis is an important post-translational mechanism that underlies irreversible regulation of protein functions. More specifically, the profile helps to exactly determine limited cleavage sites for signal peptide or propeptide removal, peptide hormone processing, ectodomain shedding, and regulated intramembrane proteolysis (Figure). Incorrectly predicted signal cleavage sites are found in major proteins such as extracellular matrices, secreted enzymes and the peptide hormone precursor adrenomedullin (J Proteome Res 14: 4921-31, 2015). This study demonstrates the utility and feasibility of peptidomics in biochemical research of a given protein.

last updated:2021/10/01
